New Features in MLlib in Apache Spark 1.0

MLlib is an Apache Spark component focusing on machine learning. It became a standard component of Spark in version 0.8 (Sep 2013). The initial contribution was from Berkeley AMPLab. Since then, 50+ developers from the open source community have contributed to its codebase. With the release of Apache Spark 1.0, I’m glad to share some of the new features in MLlib. Among the most important ones are:
- sparse data support
- regression and classification trees
- distributed matrices
- PCA and SVD
- L-BFGS optimization algorithm
- new user guide and code examples
This is the first in a series of blog posts about features and optimizations in MLlib. We will focus on one feature new in 1.0 — sparse data support.
Large-scale ≈ Sparse
When I was in graduate school, I wrote “large-scale sparse least squares” in a paper draft. My advisor crossed out the word “sparse” and left a comment: “Large-scale already implies sparsity, so you don’t need to mention it twice.” It is a good argument. Sparse datasets are indeed very common in the big data world, where the sparsity may come from many sources, e.g.,
- feature transformation: one-hot encoding, interaction, and binarization,
- large feature space: n-grams,
- missing data: rating matrix.
Take the Netflix Prize qualifying dataset as an example. It contains around 100 million ratings generated by 480,189 users on 17,770 movies. Therefore, the rating matrix contains only 1% nonzeros. If an algorithm can utilize this sparsity, it can see significant improvements.
Exploiting Sparsity
In Apache Spark 1.0, MLlib adds full support for sparse data in Scala, Java, and Python (previous versions only supported it in specific algorithms like alternating least squares). It takes advantage of sparsity in both storage and computation in methods including SVM, logistic regression, Lasso, naive Bayes, k-means, and summary statistics.
To give a concrete example, we ran k-means clustering on a dataset that contains more than 12 million examples with 500 feature dimensions. There are about 600 million nonzeros and hence the density is about 10%. The result is listed in the following table:
| sparse | dense | |
|---|---|---|
| storage | 7GB | 47GB |
| time | 58s | 240s |
So, not only did we save 40GB of storage by switching to the sparse format, but we also received a 4x speedup. If your dataset is sparse, we strongly recommend you to try this feature.
Getting Started
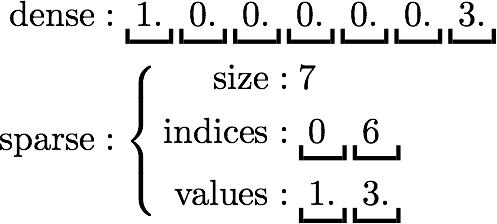
Both sparse and dense feature vectors are supported via the Vector interface. A sparse vector is represented by two parallel arrays: indices and values. Zero entries are not stored. A dense vector is backed by a double array representing its entries. For example, a vector [1., 0., 0., 0., 0., 0., 0., 3.] can be represented in the sparse format as (7, [0, 6], [1., 3.]), where 7 is the size of the vector, as illustrated below:
Take the Python API as an example. MLlib recognizes the following types as dense vectors:
- NumPy’s
array, - Python’s list, e.g.,
[1, 2, 3].
and the following as sparse vectors:
- MLlib’s
SparseVector, - SciPy’s
csc_matrixwith a single column.
We recommend using NumPy arrays over lists for efficiency, and using the factory methods implemented in Vectors to create sparse vectors.
import numpy as np
import scipy.sparse as sps
from pyspark.mllib.linalg import Vectors
# Use a NumPy array as a dense vector.
dv1 = np.array([1.0, 0.0, 3.0])
# Use a Python list as a dense vector.
dv2 = [1.0, 0.0, 3.0]
# Create a SparseVector.
sv1 = Vectors.sparse(3, [0, 2], [1.0, 3.0])
# Use a single-column SciPy csc_matrix as a sparse vector.
sv2 = sps.csc_matrix((np.array([1.0, 3.0]), np.array([0, 2]),
np.array([0, 2])), shape = (3, 1))
K-means takes an RDD of vectors as input. For supervised learning, the training dataset is represented by an RDD of labeled points. A LabeledPoint contains a label and a vector, either sparse or dense. Creating a labeled point is straightforward.
from pyspark.mllib.linalg import SparseVector
from pyspark.mllib.regression import LabeledPoint
# Create a labeled point with a positive label and a dense vector.
pos = LabeledPoint(1.0, [1.0, 0.0, 3.0])
# Create a labeled point with a negative label and a sparse vector.
neg = LabeledPoint(0.0, SparseVector(3, [0, 2], [1.0, 3.0]))
MLlib also supports reading and saving labeled data in LIBSVM format. For more information on the usage, please visit the MLlib guide and code examples.
When to Exploit Sparsity
For many large-scale datasets, it is not feasible to store the data in a dense format. Nevertheless, for medium-sized data, it is natural to ask when we should switch from a dense format to sparse. In MLlib, a sparse vector requires 12nnz+4 bytes of storage, where nnz is the number of nonzeros, while a dense vector needs 8n bytes, where n is the vector size. So storage-wise, the sparse format is better than the dense format when more than 1/3 of the elements are zero. However, assuming that the data can be fit into memory in both formats, we usually need sparser data to observe a speedup, because the sparse format is not as efficient as the dense format in computation. Our experience suggests a sparsity of around 10%, while the exact switching point for the running time is indeed problem-dependent.
Sparse data support is part of Apache Spark 1.0, which is available for download right now at https://spark.apache.org/. We will cover more new features in MLlib in a series of posts. So stay tuned.

