Apache Spark as a platform for large-scale neuroscience

The brain is the most complicated organ of the body, and probably one of the most complicated structures in the universe. It’s millions of neurons somehow work together to endow organisms with the extraordinary ability to interact with the world around them. Things our brains control effortlessly -- kicking a ball, or reading and understanding this sentence -- have proven extremely hard to implement in a machine.
For a long time, our efforts were limited by experimental technology. Despite the brain having many neurons, most technologies could only monitor the activity of one, or a handful, at once. That these approaches taught us so much -- for example, that there are neurons that respond only when you look at a particular object -- is a testament to experimental ingenuity.
In the next era, however, we will be limited not by our recordings, but our ability to make sense of the data. New technologies make it possible to monitor the activity of many thousands of neurons at once -- from a small region of the mouse brain, or from the entire brain of the larval zebrafish. These advances in recording come with dramatic increases in data size. Several years ago, a large neural data set might have been a few GB, amassed across months or years. Today, monitoring the entire zebrafish brain can yield several TBs in an hour (see this paper for a description of recent experimental technology).
Whole-brain activity measured in a larval zebrafish while simultaneously presenting a moving visual stimulus (upper left) and monitoring intended swimming behavior (size of circle)
There are many challenges to analyzing neural data. The measurements are indirect, and useful signals must be extracted and transformed, in a manner tailored to each experimental technique -- our version of ETL. At a higher-level, our analyses must find patterns of biological interest from the sea of data. An analysis is only as good as the experiment it motivates; the faster we can explore data, the sooner we can generate a hypothesis and move research forward.
In the past, neuroscience analysis has largely relied on single-workstation solutions. In the future, it will increasingly rely on some form of distributed computing, and we think Spark is the ideal platform. This post explains why.
Why Apache Spark?
The first challenge in bringing distributed computing to a community not currently using it is deployment. Apache Spark can run out-of-the-box on Amazon’s EC2, which immediately opens it up to a wide community. Although the cloud has many advantages, several universities and research institutes have existing high-performance computing clusters. We were pleased and surprised by how straightforward it was to integrate Spark into our own cluster, which runs the Univa Grid Engine. We used the Spark standalone scripts, alongside the existing UGE scheduler, to enable our users to launch their own private Spark cluster with a pre-specified number of nodes. We did not need to setup Hadoop, because Spark can natively load data from our networked file system. Our infrastructure is common to many academic and research institutions, and we hope it will be easy to replicate our approach elsewhere.
One of the reasons neural data analysis is so challenging -- and so fascinating -- is that little of what we do is standardized. Unlike, say, trying to maximize accuracy of user recommendations, or performance of a classifier, we are trying to maximize our understanding. To be sure, there are families of workflows, analyses, and algorithms that we use regularly, but it’s just a toolbox, and a constantly evolving one. To understand data, we must try many analyses, look at the results, modify at many levels -- whether adjusting preprocessing parameters, or developing an entirely new algorithm -- and inspect the results again.
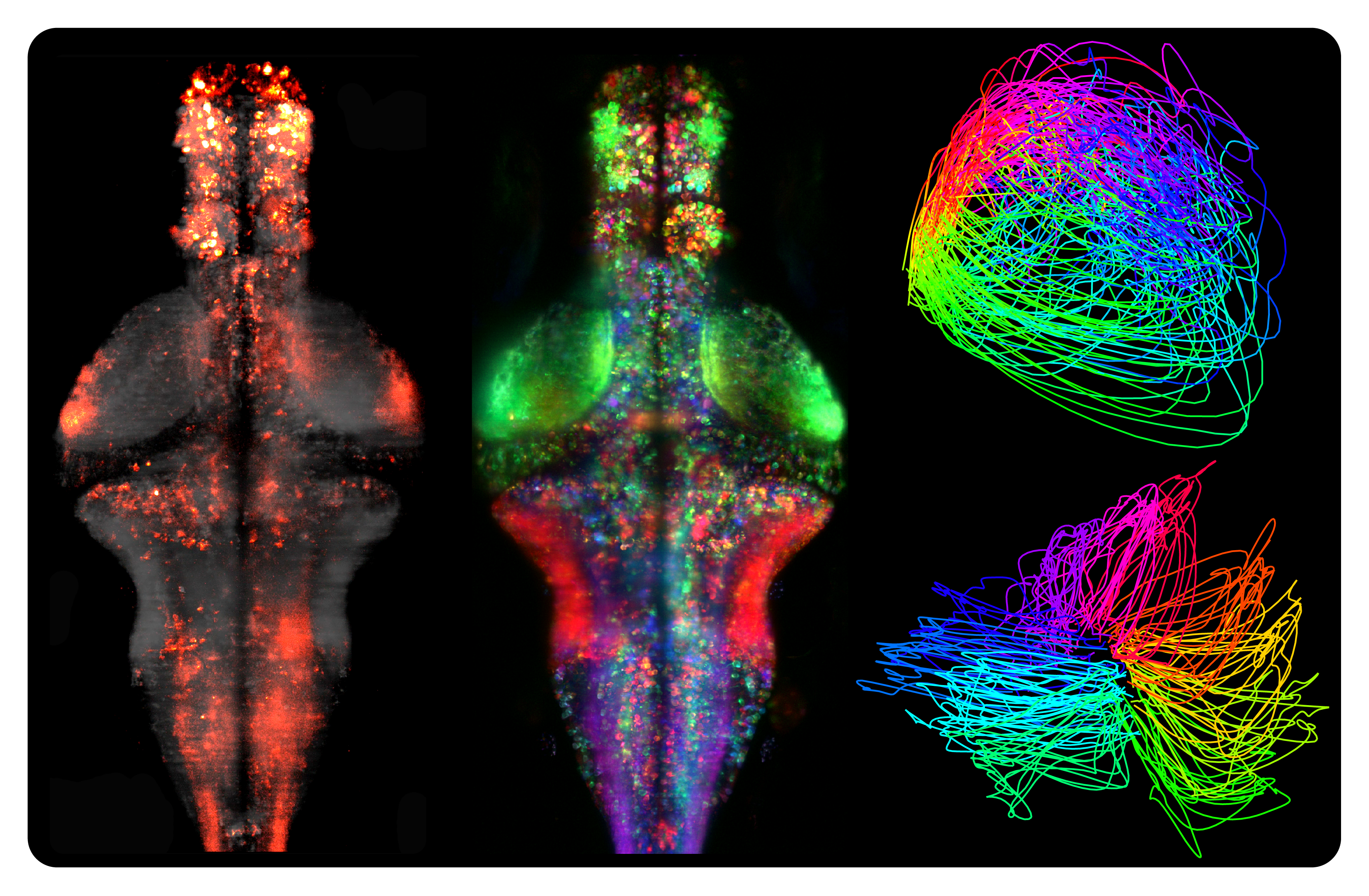
Examples of analyses performed using Spark, including basic processing of activity patterns (left), matrix factorization to characterize functionally similar regions (as depicted by different colors) (middle), and embedding dynamics of whole-brain activity into lower-dimensional trajectories (right)
The ability to cache a large data set in RAM and repeatedly query it with multiple analyses is critical for this exploratory process, and is a key advantage of Spark compared to conventional MapReduce systems. Any data scientist knows there is a “working memory for analysis”: if you need to wait more than a few minutes for a result, you forget what you were doing. If you need to wait overnight, you’re lost. With Spark, especially once data is cached, we can get answers to new queries in seconds or minutes, instead of hours or days. For exploratory analysis, this is a game changer.
Why PySpark?
Spark offers elegant and powerful APIs in Scala, Java, and Python. We are developing a library for neural data analysis, called Thunder, largely in the Python API (PySpark), which for us offers several unique advantages. (A paper describing this library and its applications, in collaboration with the lab of Misha Ahrens, and featuring Spark developer Josh Rosen as a co-author, was recently published in Nature Methods, and is available here.)
1) Ease of use. Although Scala has many advantages (and I personally prefer it), for most users Python is an easier language to learn and use for developing analyses. Due to its scientific computing libraries (see below), Python adoption is increasing in neuroscience and other scientific fields. And in some cases, users with existing analyses can bring them into Spark straightaway. To consider a simple example, a common workflow is to fit individual models to thousands or millions of neurons or neural signals independently. We can easily express this embarrassingly-parallel problem in PySpark (see an example). If a new user wants to do the same analysis with their own, existing model-fitting routine, already written in Python and vetted on smaller scale data, it would be plug-and-play. More exciting still, working in Spark means they can use the exact same platform to try more complex distributed operations: for example, take the parameters from those independently fitted models and perform clustering or dimensionality reduction -- all in Python.
2) Powerful libraries. With libraries like NumPy, SciPy, and scikit-learn, Python has become a powerful platform for scientific computing. When using PySpark, we can easily leverage these libraries for components of our analyses, including signal processing (e.g. fourier transforms), linear algebra, optimization, statistical computations, and more. Spark itself offers, through its MLlib library, many high-performance distributed implementations of machine learning algorithms. These implementations nicely complement the analyses we are developing. In so far as there is overlap, and our analyses are sufficiently general-purpose, we are either using -- or are in the process of contributing to -- analyses in MLLib. But much of what we do, and how we implement it, is specific to our problems and data types (e.g. images and time series). With Thunder, we hope to provide an example of how an external library can thrive on top of Spark (see also the ADAM library for genomic analysis from the AmpLab).
3) Visualization. Especially for exploratory data analysis, the ability to to visualize intermediate results is critical, and often these visualizations must be tailored and tweaked depending on the data. Again, the combination of Spark and Python offers many advantages. Python has a core plotting library, matplotlib, and new libraries are improving its aesthetics and capabilities (e.g. mpld3, seaborn). In an iPython notebook, we can perform analyses with Spark and visually inspect results (see an example). We are developing workflows in which custom visualizations are tightly integrated into each of our analyses, and consider this crucial as our analyses become increasingly complex.
The future: Spark Streaming
Spark has already massively sped up our post-hoc data processing and analysis. But what if we want an answer during an experiment? We are beginning to use Spark Streaming for real-time analysis and visualization of neural data. Because Spark Streaming is built in to the Spark ecosystem, rather than an independent platform, we can leverage a common code-base, and also the same deployment and installation. Streaming analyses will let us adapt our experiments on the fly. And as technology marches ever forward, we may soon collect data so large and so fast that we couldn’t store the complete data sets even if we wanted to. By analyzing more and more online, and storing less, Spark Streaming may give us a solution.
Conclusion
We are at the beginning of an exciting moment in large-scale neuroscience. We think Spark will be core to our analytics, but significant challenges lie ahead. Given the scale and complexity of our problems, different research groups must work together to unify analysis efforts, vet alternative approaches, and share data and code. We believe that any such effort must be open-source through-and-through, and we are fully committed to building open-source solutions. We also need to work alongside the broader data science and machine learning community to develop new analytical approaches, which could in turn benefit communities far beyond neuroscience. Understanding the brain will require all of our biological and analytical creativity -- and we just might help revolutionize data science in the process.

