Glow V1.0.0, Next Generation Genome Wide Analytics

Genomics data has exploded in recent years, especially as some datasets, such as the UK Biobank, become freely available to researchers anywhere. Genomics data is leveraged for high-impact use cases – gene discovery, research and development prioritization, and to conduct randomized controlled trials. These use cases will help in developing the next generation of therapeutics.
The catch: deriving insights from this data requires data teams to scale their analytics. And scaling requires data scientists and engineers with deep technical skill sets. That’s why we’re excited to announce the release of Glow version 1.0.0, an open source library that solves key challenges of applying distributed computing to genomics data in the cloud.
Challenges with genetic association studies
As genetic data has grown, processing, storing and analyzing it has become a major bottleneck. Challenges include:
- Variety of data. The variety of data types can make managing it a real headache. For instance, Biobank data contains genomics, electronic health records, medical devices and images.
- Volume and velocity of data. Genetic data is massive and constantly evolving, and analyses are rerun continually as fresh data comes in.
- Inflexible analytics. Single node bioinformatics tools do not allow users to work together interactively on large datasets. Genomics data formats may be optimized for compression and storage, but not for analytics. Bioinformatics scientists filter samples that are either from the same family or of different ethnicities. Hard filtering limits the power to make new discoveries.
Introducing Glow
Glow is an open-source toolkit for working with genomic data at population-level scale. The toolkit is natively built on Apache Spark™, a unified analytics engine for large-scale data processing and machine learning.
- Bridges bioinformatics and the big data ecosystem. With Glow, you can ingest variant call format (VCF), bgen, plink and Hail matrix tables under a common variant schema. Variant data can then be written to Delta Lake to create genomics data lakes, which can be linked to a variety of data sources using distributed machine learning algorithms such as GraphFrames.
- Built to scale. Glow natively builds on Apache Spark™ and Delta Lake, allowing users to ramp from 1 to 10 to 100 nodes. Scaling computers is faster than optimizing code or hardware.
- Natively supports genetic association studies.Glow is concordant with regenie for linear and logistic regression and now supports up to 20 phenotypes simultaneously. The method allows you to include all the data without filtering, and controls for an imbalance of cases and controls. Glow is written using Python and Pandas user defined functions, allowing computational biologists to extend Glow to gene burden or joint variant analysis, for example.
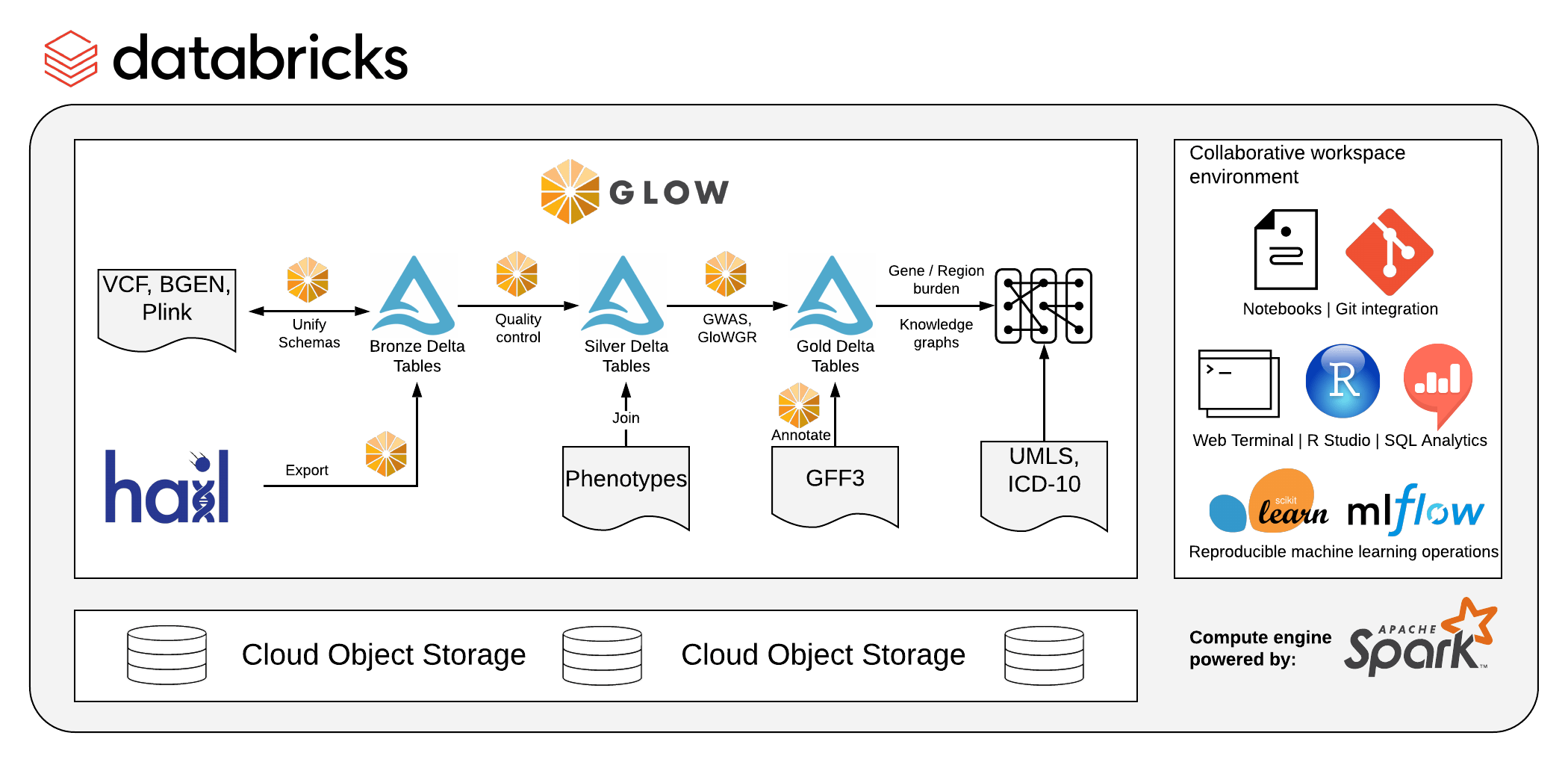
Figure 1. The Glow library can be run on Databricks on any of the three major clouds, starter notebooks can be found on the documentation.
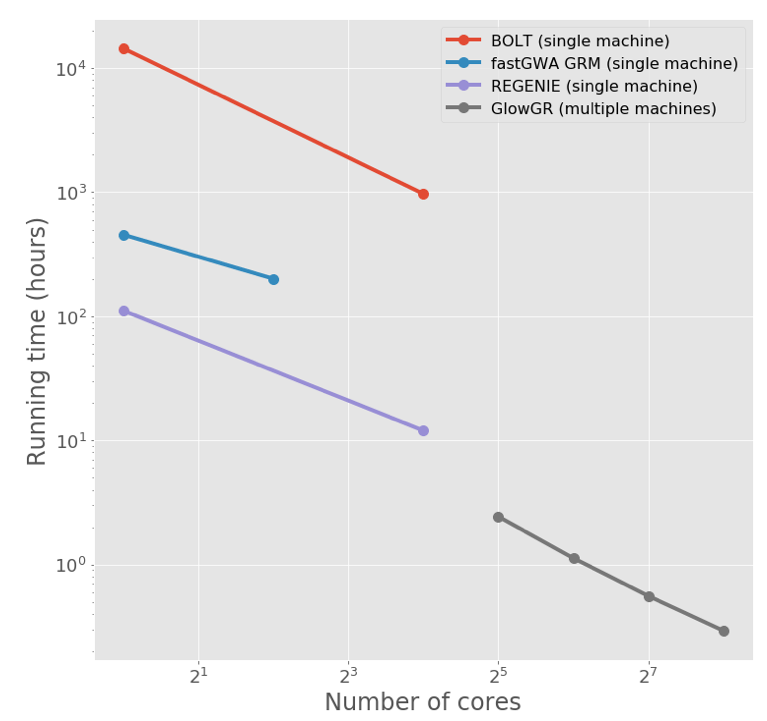
Figure 2. Glow’s whole genome regression (GloWGR) is orders of magnitude more scalable than existing methods
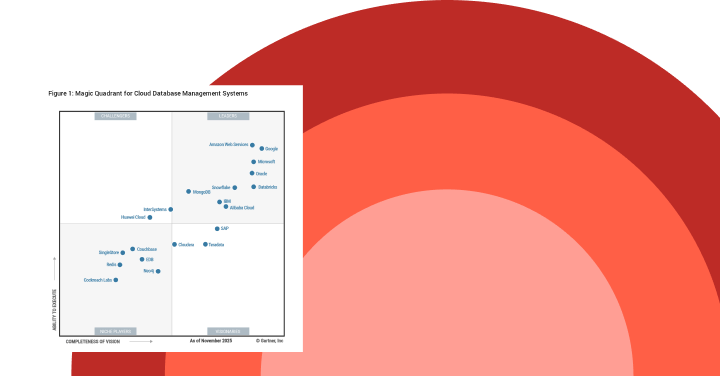
Gartner®: Databricks Cloud Database Leader
Conclusions
We have collaborated with the Regeneron Genetics Center to solve key scaling challenges in genomics through project Glow. Bioinformatics, computational biologists, statistical geneticists and research scientists can work together on The Databricks analytics platform, on any cloud, to scale their genomics data analytics and downstream machine learning applications. The first use case of Apache Spark™ and Delta Lake to genomics has been for population genetic association studies. And we are now seeing new use cases emerging for cancer and childhood developmental disorders.
Get started
Try out Glow V1.0.0 on Databricks or learn more at projectglow.io.