Extracting Oncology Insights From Real-World Clinical Data With NLP
Preview the solution accelerator notebooks referenced in this blog online or get started right away by downloading and importing the notebooks into your Databricks account.
Cancer is the leading cause of death and disease in the U.S., and the numbers are staggering with nearly 2 million new cases of cancer expected to be diagnosed in the U.S. this coming year. Cancer also represents a significant portion of total U.S. healthcare spending, estimated at more than $200B in 2020. As such, the biopharmaceutical industry is heavily focused on oncology drug development. Nearly 40 new cancer drugs were approved by the FDA in 2019 and 2020 alone, and more than 1300 new medications and vaccines are in clinical development.
Measuring the efficacy of oncology interventions is critical to matching patients with the right intervention. Oncology data, and related real-world evidence, have the potential to inform clinical research, trial design, regulatory decisions, safety assessments, treatment pathways and more. Unfortunately, given the highly specialized nature of oncology care, disease criteria and endpoints typically are not available in structured formats and remain locked in data silos, making them hard to aggregate and analyze.
In oncology, pathology reports (often captured in PDF format and siloed in EMR systems), contain critical information, such as tumor size, grade, stage and histology. These variables, once extracted with a natural language processing (NLP) system, can be used to define disease cohorts, assess disease severity and create a baseline for disease progression, which then can be applied to the aforementioned use cases, ranging from clinical trial matching to treatment pathways. But extracting this information from unstructured clinical text data is often a huge pain point for data teams.
John Snow Labs, the leader in healthcare NLP, and Databricks are tackling these challenges head-on and working with many customers across the healthcare ecosystem to translate unstructured oncology data into actionable evidence.
Clinical natural language processing at scale with Databricks & John Snow Labs
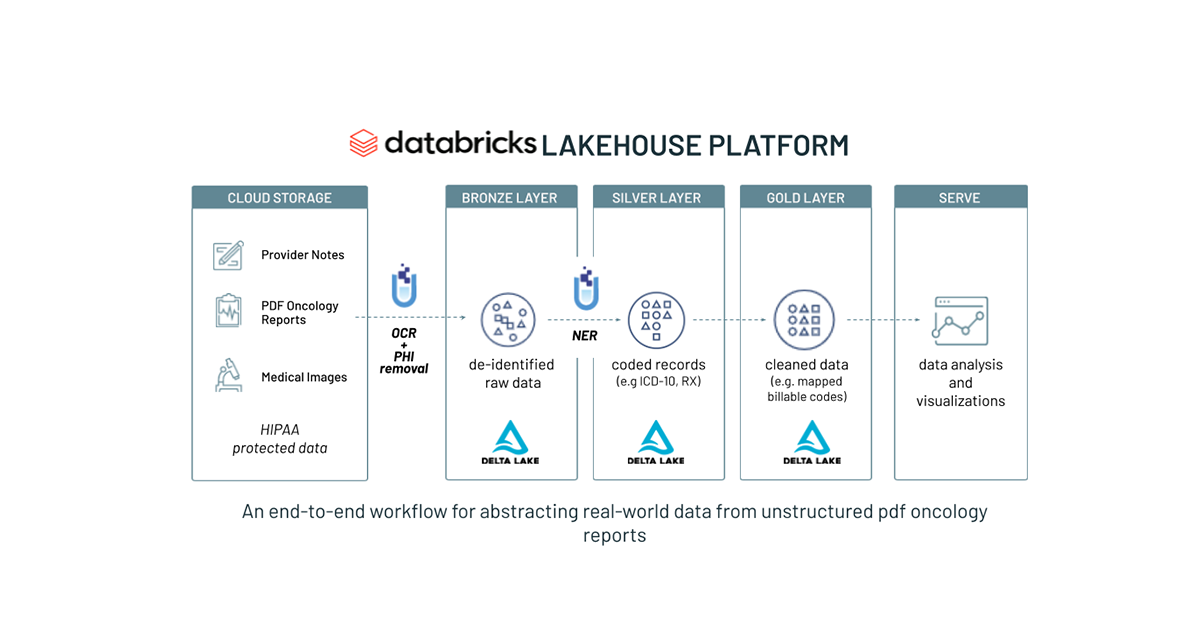
The path forward begins with the Databricks Lakehouse Platform, a modern data platform that combines the best elements of a data warehouse—such as data management and performance —with the low cost, flexibility and scale of a cloud data lake. This new, simplified architecture enables health systems to unify all their data—structured (e.g. diagnoses and procedure codes found in EHR databases), semi-structured (e.g. HL7, FHIR messages) and unstructured (e.g. free-text notes and images)— into a single, high-performance platform for both traditional analytics and data science.
At the core of the Databricks Lakehouse Platform is Delta Lake, an open-source storage layer that brings performance (via Apache Spark™), reliability and governance to a data lake. Healthcare organizations can land all of their data – including raw provider notes, radiology reports and PDF pathology reports – into Delta Lake. This preserves the original source of truth before applying any data transformations. By contrast, with a traditional data warehouse, transformations occur prior to loading the data, which means that all structured variables extracted from unstructured text are disconnected from the native text.
Building on this foundation is John Snow Labs' Spark NLP for Healthcare, the most widely-used NLP library in the healthcare and life science industries. Optimized to run on Databricks, Spark NLP for Healthcare seamlessly extracts, classifies and structures clinical and biomedical text data with state-of-the-art accuracy at scale. It is the only native distributed open-source text processing library for Python, Java and Scala, and since every Spark NLP pipeline is a Spark ML pipeline, it is particularly well suited to building unified NLP and machine learning pipelines. Spark NLP provides Python, Java and Scala libraries with the full functionality of traditional NLP libraries (like spaCy, nltk, Stanford CoreNLP and Open NLP) and adds additional functionality, such as spell-checking, sentiment analysis and document classification. You can learn more about the joint Databricks and John Snow Labs solution in our previous blog, Applying Natural Language Processing to Health Text at Scale.
Get started with ETL
Real-world oncology data abstraction in action
To demonstrate the power of Databricks and John Snow Labs, we created a Solution Accelerator for abstracting real-world data from oncology notes. The solution accelerator contains sample data, prebuilt code and step-by-step instructions for ingesting and preparing oncology reports for downstream analytics and real-world evidence generation. The solution is ready to go in a Databricks notebook and to help you get started, we've included a brief walkthrough of the solution below.
For this solution we used the MT ONCOLOGY NOTES dataset. It offers resources primarily in the form of transcribed sample medical reports across medical specialties and common medical transcription words/phrases encountered in specific sections that form part of a medical report – sections such as physical examination or PE, review of systems or ROS, laboratory data and mental status exam, among others.
We chose 50 de-identified oncology reports from the MT Oncology notes dataset as the source of the unstructured text and landed the raw text data into the Delta Lake bronze layer. For demonstration purposes, we limited the number of samples to 50, but the framework presented in this solution accelerator can be scaled to accommodate millions of clinical notes and text files.
The first step in our accelerator is to extract variables using various models for Named-Entity Recognition (NER). To do that, we first set up our NLP pipeline, which contains annotators such as documentAssembler and sentenceDetector and tokenizer that are trained specifically for healthcare-related NER. In the example below, we combined bionlp_ner, which is a clinical NER model, and jsl_ner, which is a pre-trained deep NER model for clinical terminology. We see that the mesothelioma patient is experiencing symptoms such as coughing.
Extracting named entities from texts is a great example of AI-assisted ETL: pre-trained deep learning (DL) models enable us to transform unstructured data into a structured format that can be used for downstream clinical analysis.
Once we have the symptoms extracted, we can map to ICD-10 codes, which can be used for coding automation and improving Hierarchical Condition Category (HCC) coding accuracy for Medicare Risk Adjustment. We can further use this data to analyze treatment patterns and analyze the association between symptoms and oncological entities.
We can also generate a chart to study the assertion status of these symptoms as being present, absent or associated with someone else (for example, a family member).
Continuing with the same note set, we run descriptive and visual statistics to display the most common oncology entities (example below) stratified by their assertion status.
Next, we can look at treatments, including drug frequency and duration, which form the basis of oncology regimens. Below is a screenshot of the NLP model included in our solution notebook extracting drug treatment and duration information.
We can then associate symptoms in relation to treatments, as well as disease statuses such as relapse, with confidence scores.
This data is critical for ensuring both the quality of individual patient care and population-level research, which can help determine the efficacy and safety of interventions in the real world.
Using the Databricks Lakehouse Platform, we can also easily create a database of conditions, symptoms and procedures, along with other relevant extracted information from the unstructured notes, which can then be used for downstream analysis, clinical decision support and research.
With this solution accelerator, Databricks and John Snow Labs have opened the door to extract oncology data at scale with the quality required for real-world evidence generation.
Get started extracting RWD from oncology notes with NLP
To use this solution, preview the notebooks online or get started right away by downloading and importing the notebooks into your Databricks account. The notebooks include guidance for installing the related John Snow Labs NLP libraries and license keys.
You can also visit our industry pages to learn more about our Healthcare and Life Sciences solutions.
Never miss a Databricks post
What's next?

Partners
March 7, 2024/6 min read
Databricks Expands Brickbuilder Program to Include Unity Catalog Accelerators

Data Engineering
September 4, 2024/8 min read
