Shiny and Environments for R Notebooks

At Databricks, we want the Lakehouse ecosystem widely accessible to all data practitioners, and R is a great interface language for this purpose because of its rich ecosystem of open source packages and broad use as a computing language for many non-computing scientific disciplines.
The product team at Databricks actively engages with R users to identify pain points and areas for improvement. A highly requested feature is the ability to launch and share shiny applications inside the Databricks notebooks. Previously, users could develop Shiny apps inside a hosted RStudio server on Databricks, but a key limitation was not being able to share the app URL with other users.
In addition, we consistently heard that the existing package management for R code on Databricks, a feature that was introduced in 2017, was not adequate. Users want to simply call the familiar install.packages() function inside a notebook and have the package available on all the workers of the cluster. In addition, the library isolation that was introduced for Python notebooks was attractive for R users.
Shiny inside R Notebooks
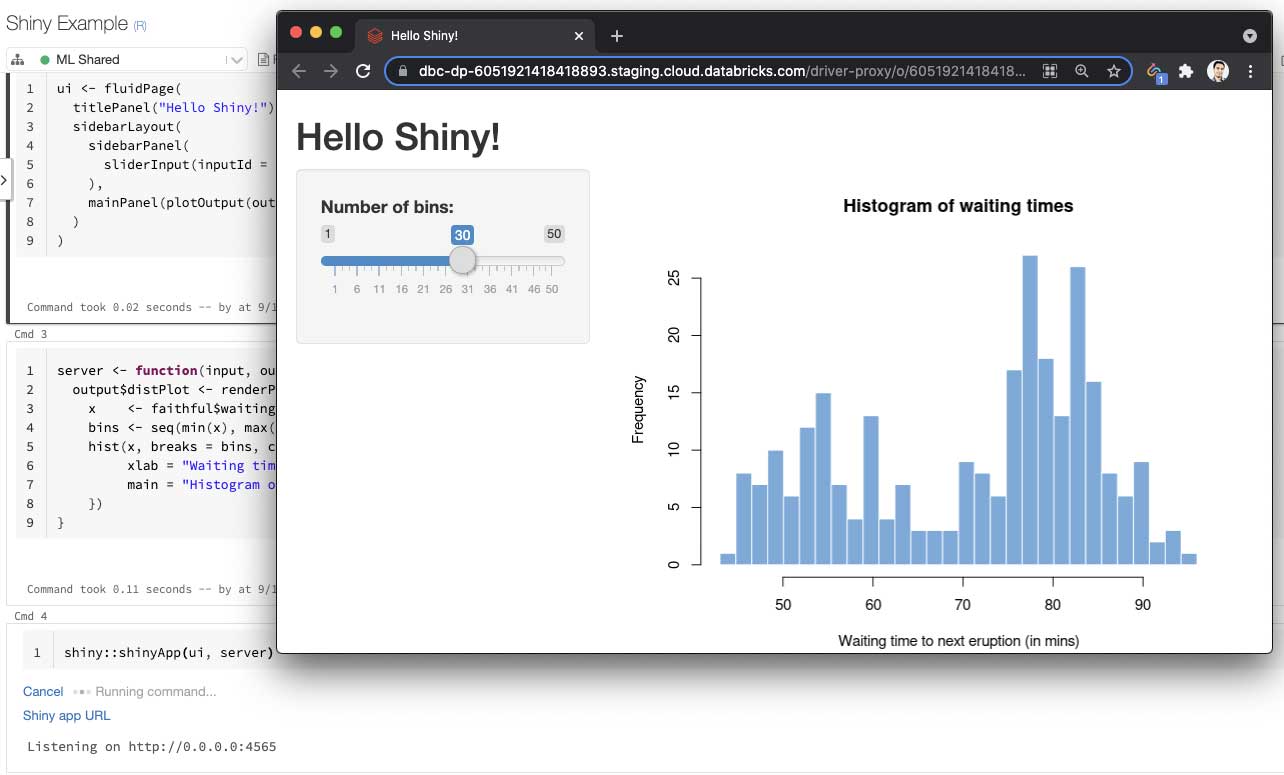
Databricks users have been able to interactively develop and test Shiny applications inside the hosted RStudio Server on Databricks. We are taking our support for Shiny to the next level by enabling R notebook users to develop and share live Shiny apps and dashboards.
Using interactive notebooks to build data applications, such as Shiny apps, is an emerging paradigm. Both notebooks and data applications are powerful tools, and data scientists naturally want to use them together. More importantly, a data application running inside a hosted notebook can be easily shared. Users would not need to “publish” Shiny applications to share them. They can simply copy the URL of the app and send it to collaborators. As long as the notebook is attached to the cluster and users have “Can Attach To” permission on the cluster, they can view and interact with the Shiny app.
To try this feature, copy the code for any sample Shiny application into a new R notebook and attach it to a cluster (or single-node) running Databricks Runtime 8.3 or above. The cell that starts the app will display a clickable link, which will open your application in a new tab.
You can use the new Databricks Repositories to check out a Shiny application from a git repository. Simply place the runApp() call in a notebook cell and launch the application.
Get started with ETL
Streaming output
Another new improvement in Databricks R notebooks is streaming standard out results of long-running commands. If you run long-running functions that print out intermediate results (e.g., iteratively optimization), you can now see the results as they are being generated. Similarly, if a piece of code generates warning messages before returning, you can view those messages in real time. This means your shiny application’s log messages will be printed in results section of the notebook cell that started the app.
Notebook-scoped libraries for R
Previously, all R notebooks running on a Databricks cluster installed packages to a single directory on the driver. This presented two limitations. First, this directory was not shared with the worker nodes, meaning that any library installed on the driver would not be accessible to Apache Spark™ workers. Second, because all notebooks installed the libraries on a shared path, users could run into version conflicts when attempting to install different versions of a package. This is surprisingly common due to transient dependencies.
With notebook-scoped libraries introduced in Databricks Runtime 9.0, each R notebook gets a unique path to install and load libraries. This path exists on a cluster-scoped NFS mount, which allows Spark workers to access libraries installed on the driver by the notebook user. This improves productivity because, as an R user, you do not need to switch out of your notebook to configure a cluster-scoped library -- simply install it inside your notebook. This gives you much better isolation on shared clusters.
When the notebook is detached, the system cleans up the per-notebook unique directory. As a result, when a notebook is detached and reattached to a cluster, a new notebook-scoped directory is generated, and the previous library state is reset.
If you are interested in understanding how R manages the package installation and import, as well as how Databricks implements notebook-scoped libraries for R notebooks, please read our user guide.
It is worth noting another approach to isolating R environments named renv. renv is an R package that lets users manage R dependencies specific to a notebook. To learn more about how to use renv inside Databricks notebooks, visit our guide for renv.