Processing Uncommon File Formats at Scale with MapInPandas and Delta Live Tables
An assortment of file formats
In the world of modern data engineering, the Databricks Lakehouse Platform simplifies the process of building reliable streaming and batch data pipelines. However, handling obscure or less common file formats still poses challenges for ingesting data into the Lakehouse. Upstream teams responsible for providing data make decisions on how to store and transmit it, resulting in variations in standards across organizations. For instance, data engineers must sometimes work with CSVs where schemas are open to interpretation, or files where the filenames lack extensions, or where proprietary formats require custom readers. Sometimes, simply requesting "Can I get this data in Parquet instead?" solves the problem, while other times a more creative approach is necessary to construct a performant pipeline.
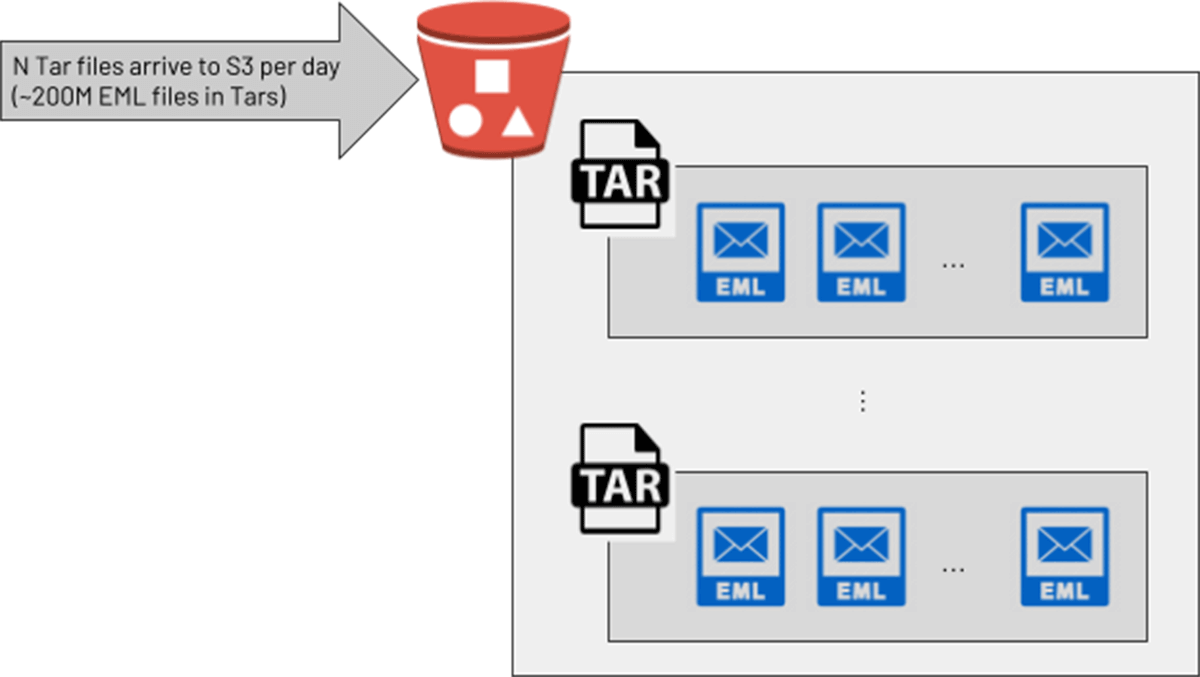
One data engineering team at a large customer wanted to process the raw text of emails for cyber security use cases on Databricks. An upstream team provided these in zipped/compressed Tar files, where each Tar contained many email (.eml) files. In the customer's development environment, engineers devised a suitable solution: a PySpark UDF invoked the Python "tarfile" library to convert each Tar into an array of strings, then used the native PySpark explode() function to return a new row for each email in the array. This seemed to be a solution in a testing environment, but when they moved to production with much larger Tar files (up to 300Mb of email files before Tarring), the pipeline started causing cluster crashes due to out-of-memory errors. With a production target of processing 200 million emails per day, a more scalable solution was required.
MapInPandas() to handle any file format
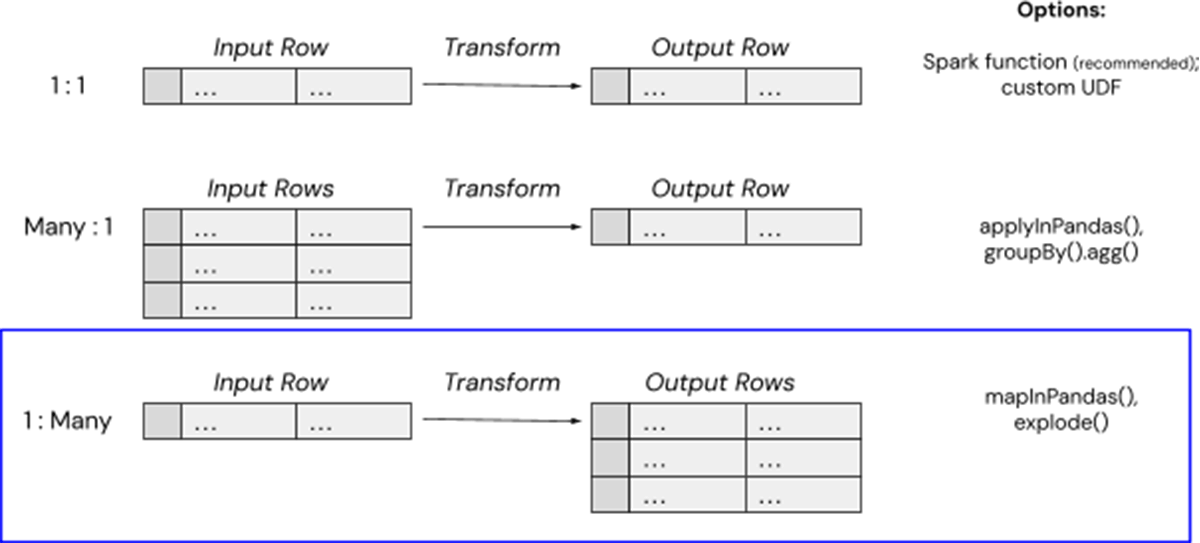
There are a few simple methods for handling complex data transformations in Databricks, and in this case, we can use mapInPandas() to map a single input row (e.g. a cloud storage path of a large Tar file) to multiple output rows (e.g. the contents of individual .eml text files). Introduced in Spark 3.0.0., mapInPandas() allows you to efficiently complete arbitrary actions on each row of a Spark DataFrame with a Python-native function and yield more than one return row. This is exactly what this high-tech customer needed to "unpack" their compressed files into multiple usable rows containing the contents of each email, while avoiding the memory overhead from Spark UDFs.
Data intelligence reshapes industries
mapInPandas() for File Unpacking
Now that we have the basics, let's see how this customer applied this to their scenario. The diagram below serves as a conceptual model of the architectural steps involved:
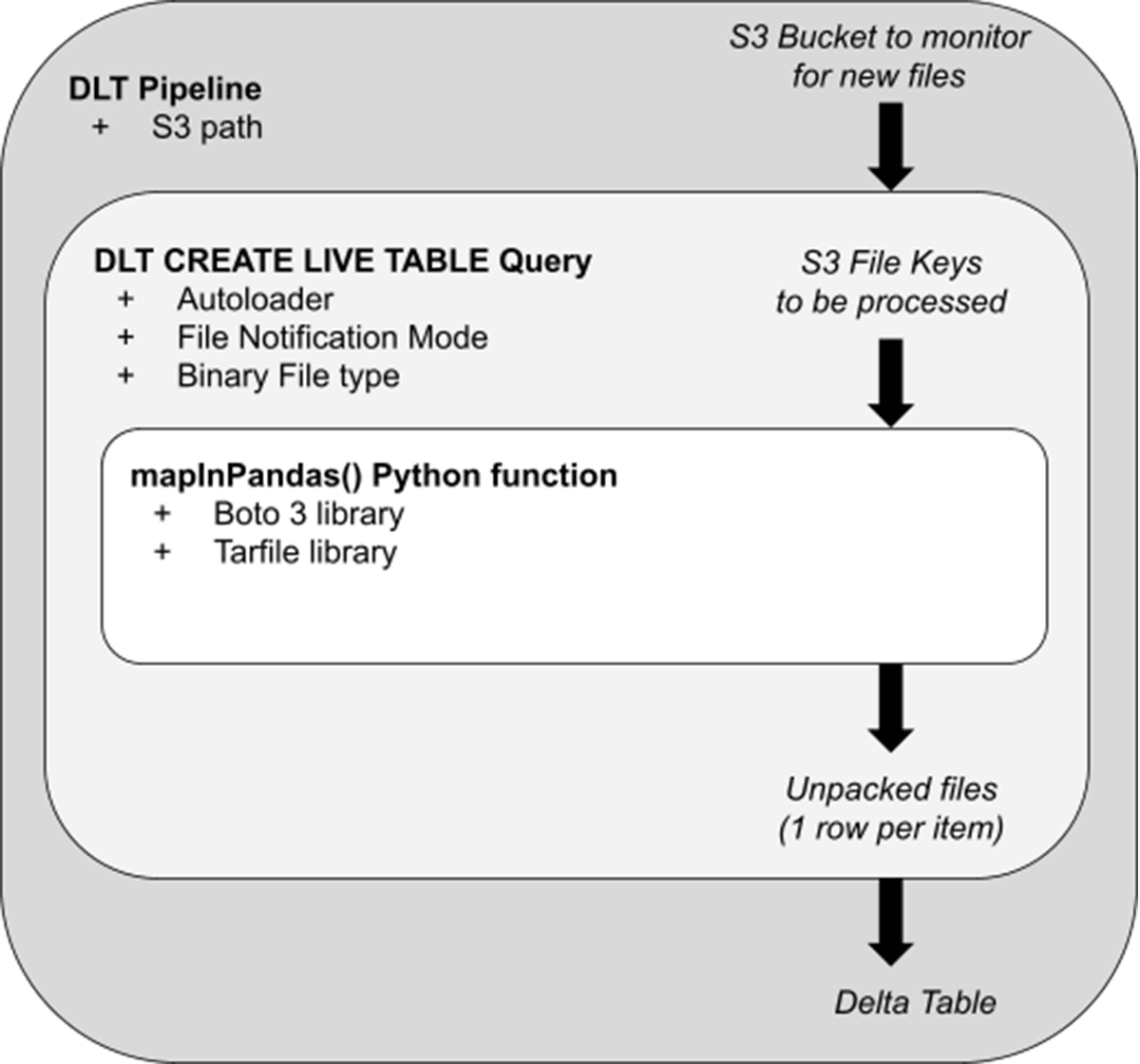
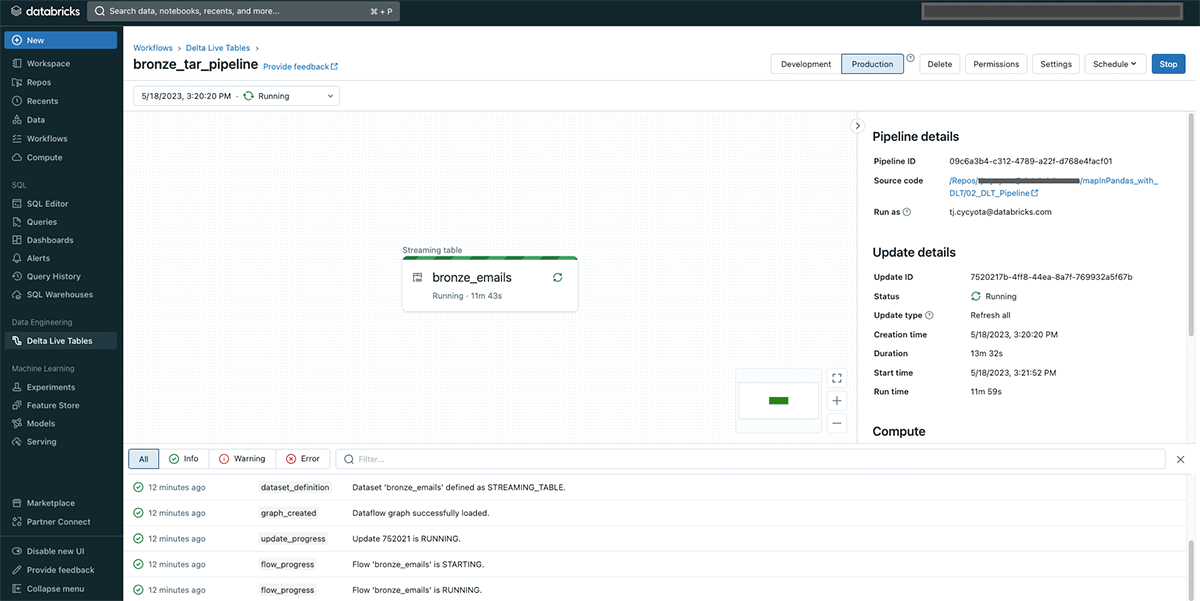
- A Delta Live Tables (DLT) Pipeline serves as the orchestration layer for our unpacking and other logic. When in Production mode, this streaming pipeline will pick up and unpack new Tar files as they arrive on S3. In preliminary testing on a non-Photon pipeline, with default DLT cluster settings, Tar files up to 430Mb were quickly processed (<30 seconds per batch) without putting memory pressure on the cluster. With enhanced autoscaling, the DLT cluster will scale up and down to match the incoming file volume, as each worker is executing the unpacking in parallel.
- Within the pipeline, a "CREATE STREAMING TABLE" query specifies the S3 path from which the pipeline ingests. With File Notification mode, the pipeline will efficiently receive a list of new Tar files as they arrive, and pass those file "keys" to be unpacked by the innermost logic.
- Passed to the mapInPandas() function is a list of files to process in the form of an iterator of pandas DataFrames. Using the standard Boto3 library and a tar-specific Python processing library (Tarfile), we'll unpack each file and yield one return row for every raw email.
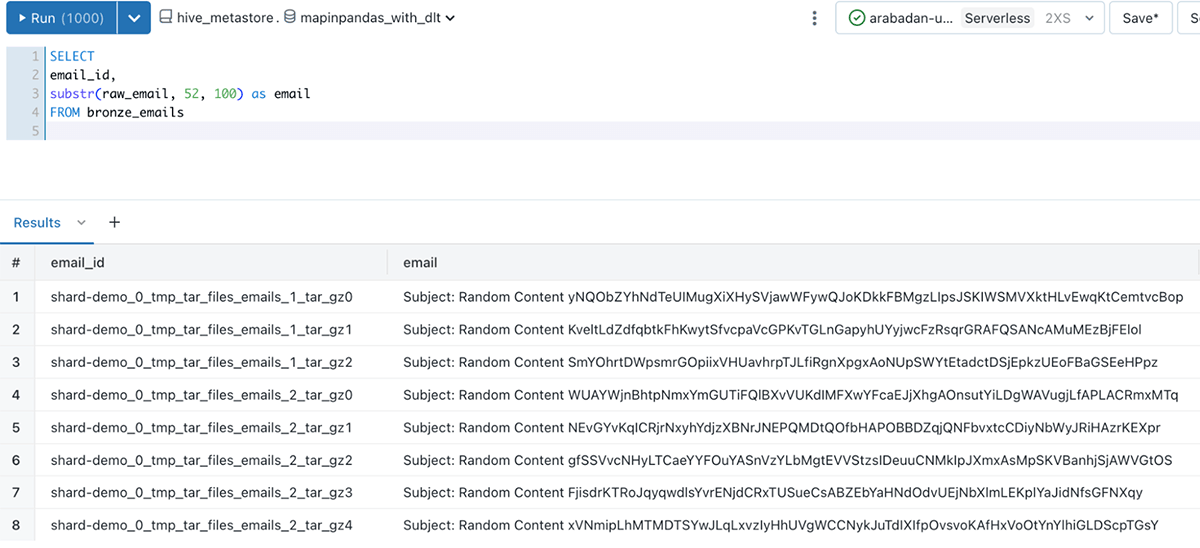
The end result is a analysis-ready Delta table that is queryable from Databricks SQL or a notebook that contains our email data, and the email_id column to uniquely identify each unpacked email:
The notebooks showcasing this solution contain the full mapInPandas() logic, as well as pipeline configuration settings. See them here.
Further Applications
With the approach described here, we have a scalable solution to process Tar email files at low latency for important business applications. Delta Live Tables can be quickly adjusted to match file arrival volumes, as we can switch a pipeline from continuous to triggered without any changes to the underlying code. While this example focused on the "bronze" layer of ingesting raw files from S3, this pipeline can be easily extended with cleansing, enrichment, and aggregation steps to make this valuable data source available to business users and machine learning applications.
More generally though, this mapInPandas() approach works well for any file-processing tasks that are otherwise challenging with Spark:
- Ingesting files without a codec/format supported in Spark
- Processing files without a filetype in the filename: if
file123is actually a file of type "tar", but was saved without a .tar.gz file extension - Processing files with proprietary or niche extensions, such as the Zstandard compression algorithm: simply replace the innermost loop of the MapInPandas function with the Python library needed to emit rows.
- Breaking down large, monolithic, or inefficiently stored files into DataFrame rows without running out of memory.
Find more examples of Delta Live Tables notebooks here, or see how customers are using DLT in production here.
Never miss a Databricks post
What's next?

Data Science and ML
October 1, 2024/5 min read
From Generalists to Specialists: The Evolution of AI Systems toward Compound AI
Product
November 27, 2024/6 min read