Automating Radiology Workflow with Large Language Models on Databricks
Radiology is an important component of diagnosing and treating disease through medical imaging procedures such as X-rays, computed tomography (CT), magnetic resonance imaging (MRI), nuclear medicine, positron emission tomography (PET) and ultrasound. The typical radiology workflow involves manual steps, particularly around the protocoling process. With Large Language Models (LLMs), we can automate some of this administrative burden.
Current State: Radiology Workflows
To explore in more detail, let's dive into the typical radiology workflow. Initially, a patient may visit their provider, reporting lingering effects from a recent concussion. The provider compiles the patient's notes into their Electronic Health Record (EHR) and requests imaging, such as a CT scan. Subsequently, a radiologist reviews the clinical notes and assigns an appropriate protocol label to the order, such as 'CT of Brain with contrast.' This label guides the imaging technician in executing the order, leading to the exam and subsequent review of results.
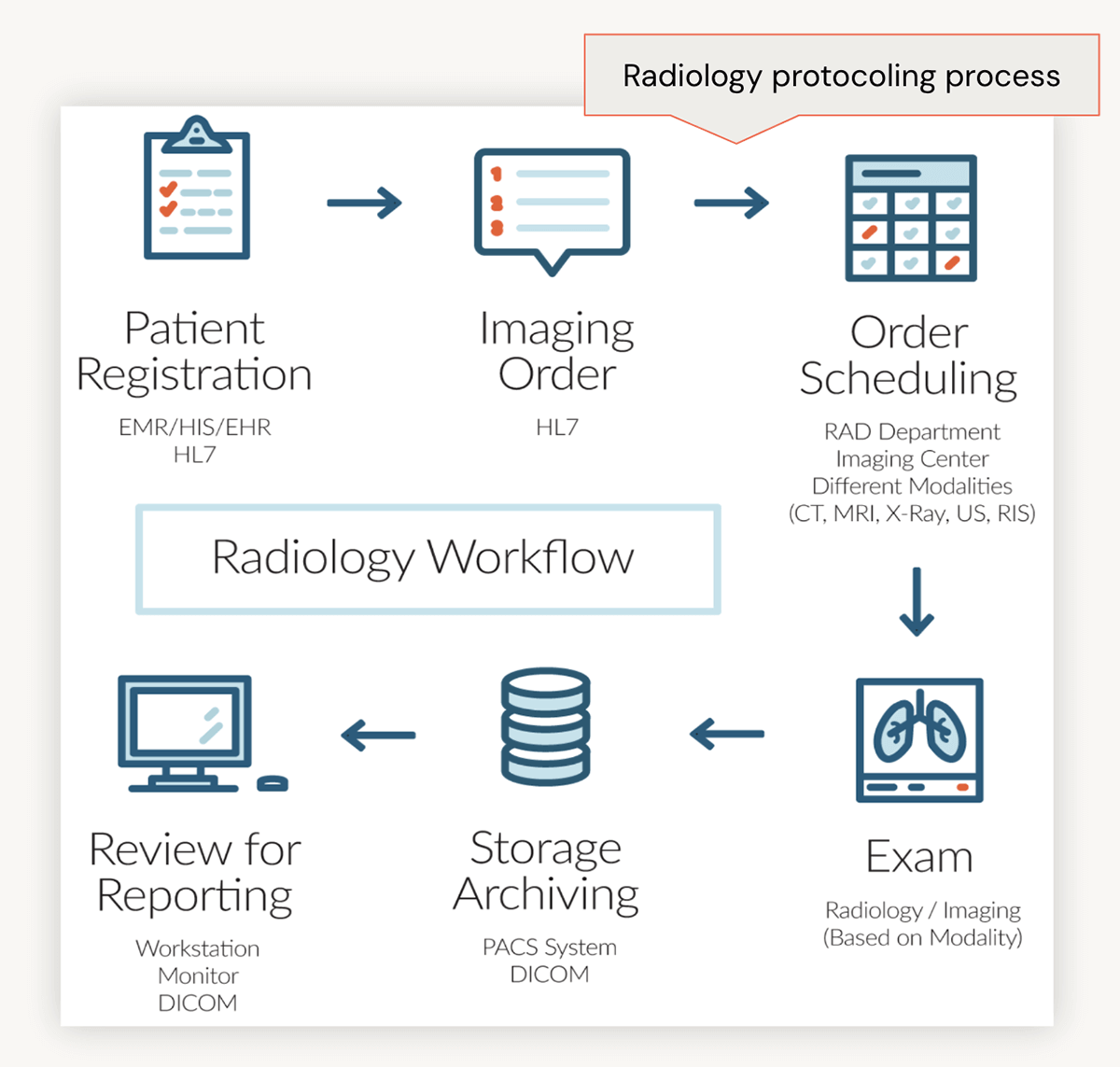
Identifying Administrative Burden in Radiology Workflows
Now, why is the manual process of assigning protocol labels significant? There are two primary reasons.
Firstly, to mitigate inaccuracies inherent in human error. Our aim isn't to replace radiologists, but to augment their decision-making process, increasing consistency and reducing errors.
Secondly, radiologists, being among the highest-paid medical professionals, spend approximately 3.5 - 6.2%(1,2) of their time, roughly equivalent to $17,000 to 30,000 annually, on label assignments. By optimizing this process, radiologists can redirect their efforts toward more impactful tasks.
Addressing Manual Tasks with Large Language Models (LLMs)
How did we address this challenge? Initially, we generated a synthetic dataset using ChatGPT, mimicking provider notes and corresponding protocol labels. While not recommended for customer use due to legal constraints, it effectively supports this proof of concept.
We then selected Meditron-7b as the foundational LLM for fine-tuning. Meditron, an open-source LLM trained on PubMed articles, is apt for healthcare and life sciences applications, and still can be improved with fine-tuning. Foundation models like Meditron are broad, and our use case demands specificity. Fine-tuning tailors the model to our specialized requirements around radiology protocoling.
To ensure cost-effectiveness, we implemented Parameter Efficient Fine Tuning (PEFT). This approach entails freezing a subset of parameters during the fine-tuning process, thereby optimizing only a fraction of parameters. Through decomposition of the parameter matrix, we markedly reduce computational requirements while preserving performance. For instance, consider a matrix (W) representing pretrained weights with dimensions of 10,000 rows by 20,000 columns, yielding a total of 200 million parameters for updating in conventional fine-tuning. With PEFT, we decompose this matrix into two smaller matrices using an inner dimension hyperparameter. For example, by decomposing the W matrix into matrices A (10,000 rows by 8 columns) and B (8 rows by 20,000 columns), we only need to update 240,000 parameters. This signifies an approximate 99% reduction in the number of parameters requiring updates!
We implemented QLoRA (Quantized Lower Rank Adaption) as part of our PEFT strategy. QLoRA is a 4-bit transformer that integrates high-precision computing with a low-precision storage approach. This ensures the model remains compact while maintaining high performance and accuracy levels.
QLoRA introduces three innovative concepts aimed at reducing memory usage while maintaining high-quality performance: 4-bit Normal Float, Double Quantization, and Paged Optimizers.
- 4-bit Normal Float: This novel data type facilitates freezing subsets of neural networks, enabling Parameter Efficient Fine Tuning (PEFT) to achieve performance levels comparable to 16-bit precision. By back-propagating gradients through a frozen 4-bit pre-trained Language and Learning Model (LLM) to LoRA, the weight matrix is decomposed based on the supplied rank parameter (r) during the fine-tuning process. Lower values of r yield smaller update matrices with fewer trainable parameters.
- Double Quantization: This process involves mapping model weights to lower bit depths to reduce memory and computational requirements. For instance, starting with a 16-bit NF, it can be quantized to 8 bits, followed by a subsequent quantization to 4 bits. This method, applied twice, results in significant memory savings, approximately 0.37 bits per parameter on average, equivalent to approximately 3GB for a 65B model (4).
- Paged Optimizers: Leveraging the concept of a "pager," which manages data transfer between main memory and secondary storage, Paged Optimizers optimize the arrangement of model weights and biases in memory. This optimization minimizes the time and resources required, particularly during the processing of mini-batches with extended sequence lengths. By preventing memory spikes associated with gradient checkpointing, Paged Optimizers mitigate out-of-memory errors, historically hindering fine-tuning large models on a single machine.
Our pipeline runs entirely on Databricks and follows a structured flow: We acquired the base Meditron model from Hugging Face into Databricks, imported our synthetic dataset into Unity Catalog (UC) Volumes, and executed the fine-tuning process with PEFT QLoRA in Databricks.
We stored model weights, checkpoints, and tokenizer in UC Volumes, logged the model with MLflow, and registered it in UC. Lastly, we utilized PySpark for inference with batch prediction. Semantic similarity served as our evaluation metric, considering context in the evaluation process and ensuring versatility across text generation and summarization tasks.
The benefit of executing this pipeline within Databricks lies in its comprehensive workflow capabilities and integrated governance across all layers. For instance, we can restrict access to sensitive patient notes authored by physicians, ensuring that only authorized personnel can view them. Similarly, we can control the usage of model outputs, ensuring that they are accessed only by relevant healthcare professionals, such as radiologists.
Moreover, it's noteworthy that all features utilized in this setup are generally available, ensuring compliance with HIPAA regulations.
We trust that our customers will discover this pipeline to be a valuable asset for optimizing memory usage across various fine-tuning scenarios in healthcare and life sciences.
Picture this: the next time a radiologist reviews a physician's notes, detailing complex medical history and diagnoses—such as 'History: heart failure, hepatic vein; Diagnosis: concern for liver laceration post-procedure, post-biopsy, on apixaban'—our system ensures the accurate assignment of the appropriate protocol label, like 'CT Abdomen W Contrast,' rather than 'CT Brain.' This precision can significantly enhance diagnostic efficiency and patient care.
Get started on your LLM use case with this Databricks Solution Accelerator.
Databricks Solution Accelerator
References
(1) Dhanoa D, Dhesi TS, Burton KR, Nicolaou S, Liang T. The Evolving Role of the Radiologist: The Vancouver Workload Utilization Evaluation Study. Journal of the American College of Radiology. 2013 Oct 1;10(10):764–9.
(2) Schemmel A, Lee M, Hanley T, Pooler BD, Kennedy T, Field A, et al. Radiology Workflow Disruptors: A Detailed Analysis. Journal of the American College of Radiology. 2016 Oct 1;13(10):1210–4.
(3) https://www.doximity.com/reports/physician-compensation-report/2020
(4) Dettmers, Tim, Artidoro Pagnoni, Ari Holtzman, and Luke Zettlemoyer. "Qlora: Efficient finetuning of quantized llms." Advances in Neural Information Processing Systems 36 (2024).
(5) https://magazine.sebastianraschka.com/p/lora-and-dora-from-scratch
Never miss a Databricks post
What's next?

Healthcare & Life Sciences
November 14, 2024/2 min read
Providence Health: Scaling ML/AI Projects with Databricks Mosaic AI
Product
November 27, 2024/6 min read