How Elsevier Labs Implemented Dictionary Annotation at Scale with Apache Spark on Databricks
Spark Summit East is just around the corner! If you haven’t registered yet, you can get tickets here with this promo code for 20% off: Databricks20
This is a guest blog from our friend at Elsevier Labs. Sujit Pal is a Technical Research Director at Elsevier Labs. His interests are Search and Natural Language Processing.
Elsevier is a provider of scientific, technical and medical (STM) information products and services. Elsevier Labs is an advanced technology R&D group within the company. Members of Labs research and create new technologies, help implement proofs of concept, educate staff and management and represent the company in technical discussions.
In this post, we will describe the machine reading pipeline Elsevier Labs is building with Apache Spark on Databricks, with special emphasis on the Dictionary Based Entity Recognizer component. In keeping with the theme of this blog, the focus would be on the Spark related aspects of the component. You can find more information about the component itself here.
Business Challenge
Elsevier publishes approximately 350,000 articles annually across 2,000 STM journals. Our experimental machine reading pipeline reads the information in these journals in order to automatically generate knowledge graphs specialized for different STM domains. These knowledge graphs could be used for tasks ranging from summarizing text to drawing new inferences about the state of our world.
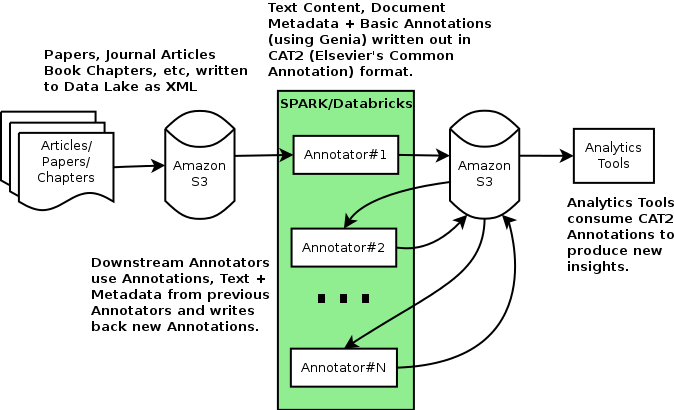
One of the major requirements for generating these knowledge graphs is to locate and identify entities mentioned in the text so they can be related to each other, and to the real-world items they name. Our machine-reading pipeline is structured as a loose sequence of batch annotators, many of which run as Databricks notebooks or jobs, where each annotator has access to the original content as well as the outputs of previous annotators in the sequence. So any solution for entity recognition needed to be accessible from the processes running in this environment.
Technology Challenge
Because of the specialized nature of STM domains, a standard off-the-shelf entity recognizer would not work for us. We did not want to spend the resources and time to build training sets that could be used to train specialized custom entity recognizers for each domain either. We decided instead to do dictionary based tagging, where we built dictionaries from readily available ontologies for different STM domains and stream our text against them. The entities that come from this step might be improved by later annotators in the pipeline in order to deal with ambiguous mentions or entities not yet in the dictionaries.
Calling external services from within Spark pipelines is generally regarded as an anti-pattern, but in this case the functionality required was quite specialized and not trivial to build into Spark. On the other hand, most of this functionality were already available in various open source components built around the Apache Solr project. So we decided to structure our dictionary based entity recognizer as a micro-service built around those components.
Our team was already using Databricks extensively, so it made sense to build our entity recognizer so it could be accessed as a micro-service from Databricks notebooks using JSON over HTTP. The interactive nature of Databricks notebooks made the process of developing and testing the service very efficient. We built a pythonic Scala API to interact with the service from the Databricks notebooks environment, both to load ontologies and to annotate text against them. The service is called using a python-style Scala API that is implemented as a custom JAR - once again, Databricks’ built in functionality to support custom JARs makes this process extremely simple.
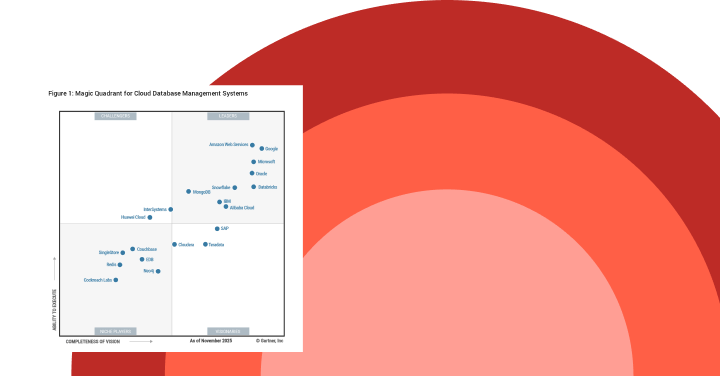
Gartner®: Databricks Cloud Database Leader
Solution
The dictionary based entity recognizer needs one or more dictionaries to compare text against. Entities are extracted from ontologies, data for which is usually available in various standard formats such as JSON-LD, RDF, etc. For each entity, a record is created consisting of a unique ID, the ontology name, and a set of synonyms. Each record is written into the Solr index.
Databricks notebooks were used to parse the data for these ontologies into the standard format as well as to load the data into the index.
The code snippet below illustrates how to load content from a flat file into the index. Each line in the input represents a single entity. Note the use of mapPartitions to instantiate the client once per partition, and the use of zipWithIndex on the inner iterator to periodically commit to the index. The last commit from the driver takes care of any straggler entities that are not yet committed.
val loaded = input.mapPartitions(p => {
val sodaClient = new SodaClient()
p.zipWithIndex.map(recIdx => {
val params = Map(
"id" -> recIdx._1._1,
"names" -> recIdx._1._2.split("::").toList,
"lexicon" -> "mesh",
"commit" -> (if (recIdx._2 % 100 == 0) true else false)
)
val req = SodaUtils.jsonBuild(params)
val resp = sodaClient.post("http://host:port/soda/add.json", req)
(recIdx._1._1, resp)
})
})
loaded.count()
val creq = SodaUtils.jsonBuild(Map("lexicon" -> "mesh", "commit" -> true))
sodaClient.post("http://host:port/soda/add.json", creq)
At query time, the client calls the microservice from a Databricks notebook, specifying a block of text (corresponding to a journal article, for example), the ontology to look up and the degree of matching required. When the client needs to annotate a large batch of documents against the same ontology, the Databricks notebook is run as a job.
For each annotation request, the microservice finds all the strings in the text that match any of the synonyms in the dictionary. The result is an RDD of entities – each entity record contains the entity ID, the text that was matched, the starting and ending character offsets of the match within the text, and a confidence measure between 0 and 1.
This RDD is then transformed into our Common Annotation (CAT2) format, using standard RDD operations on Databricks notebooks.
import com.elsevier.soda._
val resps = input.mapPartitions(p => {
val sodaClient = new SodaClient()
p.map(rec => {
val params = Map(
"lexicon" -> "countries",
"text" -> rec._2,
"matching" -> "lower"
)
val req = SodaUtils.jsonBuild(params)
val resp = sodaClient.post("http://host:port/soda/annot.json", req)
val annots = SodaUtils.jsonParseList(resp)
(rec._1, annots)
})
})
resps.count
Matching can be exact (case-sensitive or insensitive) or fuzzy. Exact matching uses the functionality provided by SolrTextTagger to stream the entire text against a Finite State Automaton created from the index. This is a very efficient operation, even for large multi-million term dictionaries. The fuzzy matching works a little differently, where we use OpenNLP to chunk the incoming text into phrases and then match against pre-normalized entity names in the index.
In addition to the annotation function just described, the microservice offers several ancillary functions to load, delete and list dictionaries, find coverage for a document across multiple dictionaries, etc.
Implementation Details
The micro-services architecture allows us to take advantage of functionality that already exists in Solr and OpenNLP. The architecture also allows us to tune the application independent of Spark. It is also horizontally scalable - by simply increasing the number of SoDA/Solr stacks and putting them behind a load balancer, we were able to achieve a linear increase in throughput.
While the loose coupling has the benefits described above, it can also lead to timeouts and job failures on the Spark side if SoDA is not able to return results in time. So it is important to make sure that SoDA (and Solr) has enough capacity to serve requests from the Spark workers within the timeout specified. On the other hand, in order to prevent idling and to get the maximum utilization out of SoDA, we need to hit it with a sufficiently large number of simultaneous requests, which translates to a suitably large number of worker threads.
Using a Databricks cluster of 1 master + 16 workers with 4 cores each, and a SoDA cluster running on 2 r3.2xlarge machines behind a load balancer, we were able to achieve a sustained annotation rate of 30 docs/sec, with an average document size of 100MB, against a dictionary of more than 8 million entries
Value Realized
Named entity recognition is critical to the success of our machine reading pipeline, and the SoDA application is one of our high-throughput strategies for achieving it. We have been using it only for a few months now, but it has already proven useful for several experiments for generating knowledge graphs from text corpora, specifically one that predicts new relationships between entities based on relationships mined from the text. Although at a basic level, the recognizer is simply an efficient solution to the problem of finding millions of named entities in millions of documents, it has the potential to help build solutions whose capabilities can extend the frontiers of scientific research.
Read the Elsevier Labs case study to learn more about how they are deploying Databricks.
Never miss a Databricks post
What's next?

Technology
December 9, 2024/6 min read
Scale Faster with Data + AI: Insights from the Databricks Unicorns Index

News
December 11, 2024/4 min read